One of the principal unsolved problems of science is the problem of protein folding, the problem of how simple strings of amino acids (called polypeptide chains) are able to form very rapidly into the intricate three-dimensional shapes that are functional protein molecules. Scientists have been struggling with this problem for more than 50 years. Protein folding is constantly going on inside the cells of your body, which are constantly synthesizing new proteins. The correct function of proteins depends on them having specific three-dimensional shapes.
In DNA, proteins are represented simply as a sequence of nucleotide base pairs that represents a linear sequence of amino acids. A series of amino acids such as this, existing merely as a wire-like length, is sometimes called a polypeptide chain.
But a protein molecule isn't shaped like a simple length of copper wire – it looks more like some intricate copper wire sculpture that some artisan might make. Below is one of the 3D shapes that protein molecules can take. There are countless different variations. Each type of protein has its own distinctive 3D shape (and some types, called metamorphic proteins, can have different 3D shapes).

The phenomenon of a polypeptide chain (a string of amino acids) forming into a functional 3D-shape protein molecule is called protein folding. How would you make an intricate 3D sculpture from a long length of copper wire? You would do a lot of folding and bending of the wire. Something similar seems to go on with protein folding, causing the one-dimensional series of amino acids in a protein to end up as a complex three-dimensional shape. In the body this happens very rapidly, in a few minutes or less. It has been estimated that it would take 1042 years for a protein to form into a shape as functional as the shape it takes, if mere trial and error were involved.
The question is: how does this happen? This is the protein folding problem that biochemists have been struggling with for decades. Recent press reports on this topic in the science press were doing what people in the science field do like crazy: making unfounded achievement boasts and parroting in a credulous fashion the boasts churned out by the PR desks of vested interests. The reports were saying that some software called AlphaFold2 (made by a company called DeepMind) had "solved" or "essentially solved" the protein folding problem. This is not at all correct.
The AlphaFold2 software gets its results by a kind of black-box "blind solution" prediction that doesn't involve real understanding of how protein folding could occur. As discussed on page 22 of this technical document, this "deep learning" occurs when the software trains on huge databases of millions of polypeptide chains (sequences of amino acids) and 3D protein shapes that arise from such sequences, databases derived from very many different organisms. One of these databases has 200 million entries, about a thousand times greater than the number of proteins in the human genome.
This is an example of what is called frequentist inference. Frequentist inference involves making guesses based on previously observed correlations. I can give an example of how such frequentist inference may involve no real understanding at all. Imagine if I had a library of vehicle images, each of which had a corresponding description such as "green van" or "red sports-car convertible" or "white tractor-trailer." I could have some fancy "deep learning" software train on many such images. The software might then be able to make a sketch that predicted pretty well what type of image would show up given a particular description such as "brown four-door station wagon." But this would be a kind of "blind solution." The software would not really know anything about what vehicles do, or how vehicles are built.
The situation I have described with this software is similar to the cases in which the Alpha Fold2 software performs well. In such cases the software uses vast external databases created from analyzing countless thousands of polypeptides sequences in countless different organisms, and 3D shapes derived from them. But it's a black box "blind solution." The software has no actual understanding of how polypeptide chains are able to form into functional 3D shapes. It cannot be true that something similar is going on inside the body. The body has no similar "deep learning" database created from crunching data on polypeptide sequences and 3D shapes derived from them, data derived from countless different organisms.
When predictions of such a type are made by deep learning software, this activity is not something corresponding to what goes when scientific theory predictions are tested against data. In the latter case what goes on is this:
(1) A theory is created postulating some way in which nature works (for example, it might be postulated that there is a universal force of attraction between bodies that acts with a certain strength, and according to a certain "inverse square" equation).
(2) Predictions are derived using the assumptions of such a theory.
(3) Such predictions are tested against reality.
But that isn't what is going on in deep-learning software. With such software, there is no theory of nature from which predictions are derived. So successes with deep-learning software aren't really explanatory science.
The type of analysis that the AlphaFold2 software does fairly well on is called template-based modeling. Template-based modeling involved frequentist inferences based on some huge database of polypeptide chains and 3D shapes that correspond to them. There's another type of way for a computer software to try to predict protein folds: what is called template-free modeling or FM. Software using template-free modeling or FM would use only the information in the polypeptide chain and the known laws of chemistry and physics, rather than relying on some huge 3D shape database (derived from many organisms) that isn't available in the human body.
How well does the Alpha Fold software do when it uses the targets that were chosen to test template-free modeling or FM? Not very well. We can find the exact data on the CASP web site.
What is called the Critical Assessment of Protein Structure Prediction (CASP) is a competition to assess the progress being made on the protein folding problem. They have been running the competition every two years since 1994. You can read about the competition and see its results at this site. The first competition in 1994 was called CASP1, and the latest competition in 2020 was called CASP14. Particular prediction programs such as AlphaFold2 are used to make predictions about the 3D shape of a protein, given a particular polypeptide chain (a sequence of amino acids specified by a gene). The competitors supposedly don't know the 3D shape, but only are given the amino acid sequence (the polypeptide sequence). The competing computer programs make their best guess about the 3D shape.
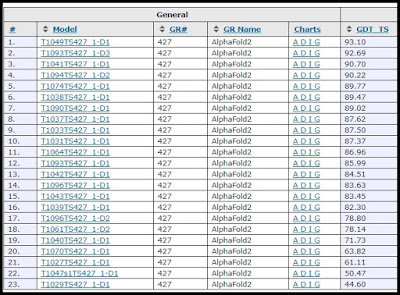
To check how well the AlphaFold2 oftware does using the targets that are supposed to test template-free modeling, you can go to the page here, click AlphaFold2 in the left box, click FM (which stands for template-Free Modeling), and then click on the Show Results button:
You will then get the results screen below:
These results are not terribly impressive. The number on the right is a number that will be close to 100 for a good prediction, and close to 0 for a very bad prediction. 19 out of 23 predictions have scored less than 90, meaning they are substantially off. A crucial consideration in judging these results is: how hard were these prediction targets? The complexity of proteins vastly differs, depending on the number of amino acids in the protein (which is sometimes referred to as the number of residues). A protein can have between 50 and 2000 different amino acids. The average number of amino acids in a human protein is about 480. If the results shown above were achieved mainly by trying to predict the 3D shapes of proteins with a below-average number of amino acids, what we can call relatively easy targets, then we should be much less impressed by the results.
What I would like to have is a column in the table above showing the number of amino acids in each prediction. The web site does not give me that. But by using the data on column 3 of this page, I can add a new column to the table above, showing the number of amino acids in the proteins that had their 3D shape predicted. Below is the result, with the new column I added on the right:
We see here that most of the predictions above were done for proteins with a smaller-than-average number of amino acids, because the numbers in the right column are mostly much smaller than 480, the average number of amino acids in a human protein. A really impressive result would be if all of the numbers in the GDT_TS column were above 90, and if most of the numbers in the last column were much larger than 480. Instead, we have a result in which AlphaFold2 often is way off (scoring much less than 90) even in simpler-than-average proteins with fewer than 300 amino acids.
Based on these results with the FM prediction targets, it is not at all true that AlphaFold2 has solved the protein folding problem. Trying to solve such targets, the software often is way off in predicting the shape of protein molecules of below-average complexity (which are easier prediction targets).
There is also a reason for thinking the not-so-great results shown in the table above would be much worse if a more reliable method was used to calculate the degree of accuracy between the prediction and the actual protein. The table above shows us numbers using a method called GDT_TS. But there's a more accurate method to calculate
the degree of accuracy between the prediction and the actual protein. That method is called GDT_HA, HA standing for "high accuracy." According to the paper here, "Of the two GDT scores under consideration, GDT_HA is generally 10–20 less than the GDT_TS scores computed from the same models (Fig 4A and 4B), reflecting its higher stringency." Figure 4 of the paper here has a graph showing that GDT_HA scores tend to be 20 or 25 points lower than GDT_TS scores. The site here also has two graphs indicating that the GDT_HA scores tend to be about 20 points lower than the GDT_TS scores. So if the more accurate GDT_HA method had been used to assess accuracy, the scores produced by AlphaFold2 for the targets picked to test template-free modeling would apparently have been only between about 24 and 73, not very impressive at all.
When we look for results using the easier targets designed to test template-based modeling (derived by using a massive "deep learning" database not available in the human body), we get results averaging about 90 for the AlphaFold2 software. But such results are using the GDT_TS scores. Based on the comment above, the more accurate GDT_HA scores would be about 20 points lower. So if the more accurate GDT_HA scores were shown we would see scores of around 70, which would not seem very impressive. Overall, judging from this page, about 75% of the CASP14 prediction targets had a number of amino acids (residues) much smaller than 480, which means that the great majority of the CASP14 prediction targets were "low-hanging fruit" that were relatively easy to solve, not a set of prediction targets that you would from get just randomly picking proteins.
Did the AlphaFold2 software stick to template-free modeling for the targets that were picked to test template-free modeling? We don't know, because the software is secret. Science is supposed to be open, available to inspection by anyone -- not some procedures hidden in secret software.
I am unable to find any claims by the DeepMind company that the AlphaFold2 software ever used any real template-free modeling based solely on the amino acids in a gene corresponding to a particular protein. On page 22 of the document here, DeepMind describes its technique, making it sound as if they used deep-learning protein database analysis for all of their predictions. So it seems as if 100% of AlphaFold2's predictions were made though what some might call the "cheat" of inappropriate information inputs, the trick of trying to predict protein folding in an organism by making massive use of information available only outside of such an organism, and derived from the analysis of proteins in very many other organisms. The fault may lie mainly with those running the CASP competitions, who have failed to enforce a sensible set of rules, preventing competitors from using inappropriate information inputs. On the current "ab initio" tab of the CASP competition, there is no mention of any requirement there ever be used any real template-free modeling based solely on the amino acids in a gene corresponding to a particular protein.
Another thing we don't know is this: how novel and secret were the prediction targets selected for the latest CASP14 competition? The targets were manually selected, and supposedly some of the prediction targets were selected in an attempt to get some novel types of prediction targets the competitors had not seen before. Were they really that, or did most of the targets tend to resemble previous year's targets, making it relatively easy for some database-based prediction program to succeed? And what type of security measures were taken to make sure that the supposedly secret prediction targets really were secret, and completely unknown to the CASP14 competitors? We don't know. Were any of the CASP14 competitors able to just find 3D shapes corresponding to some of the "secret targets," shapes that someone else had determined through molecular analysis, perhaps through some backdoor "grapevine"? We don't know. A few naughty "insider information" emails might have been sufficient for a CASP14 competitor to get information allowing it to score much higher. It's hard to predict how unlikely such a thing would be, because while we know that the military has a very good tradition of keeping secrets, we don't know how good biologists are at keeping secrets. Given very many millions of items in one of the protein databases, it would have been all-but-impossible for anyone submitting a prediction target to have known that it was something novel, not something very much like some protein already in the databases that AlphaFold2 had scanned.
It is believed that 20 to 30 percent of proteins have shapes that are not predictable from any amino acid sequence, because such proteins acheive their 3D shapes with the aid of other molecules called chaperone molecules. On such proteins a program such as AlphaFold2 would presumbably perform very poorly at its predictions. Are proteins that require chaperone molecules for their folding excluded as prediction targets for the CASP competitions, for the sake of higher prediction scores?
At this site we read "Business Insider reports that many experts in the field remain unimpressed, instead calling DeepMind’s announcement hype." There is no basis for claiming that the AlphaFold2 software has solved or "essentially solved" the protein folding problem. An example of the credulous press coverage is a news article in the journal Nature. We see a graph that is based on the template-based modeling not very relevant to human cells, since such modeling uses a massive "deep learning" database not in human cells. The graph uses the GDT_TS score, showing an average score of almost 90. We are not told that the score would have been about 20 points lower if the more accurate GDT_HA measure had been used. And we are not told that the average score using targets designed to test template-free modeling (the more relevant targets) and also using a GDT_HA measure would have been some unimpressive number such as only about 60. The graph has a misleading phrase saying, "A score above 90 is considered roughly equivalent to the experimentally determined structure." That is not correct. Since the more accurate GDT_HA measure tends to be about 20 points lower than the GDT_TS score, a prediction of a 3D protein shape can win a GDT_TS score of 90 even though it is far off the mark.
In a boastful DeepMind press release, we hear the co-founder of the CASP competition (John Moult) say this: "To see DeepMind produce a solution for this, having worked personally on this problem for so long and after so many stops and starts wondering if we’d ever get there, is a very special moment.” But in this article we read this: "AlphaFold’s predictions were poor matches to experimental structures determined by a technique called nuclear magnetic resonance spectroscopy, but this could be down to how the raw data is converted into a model, says Moult." So Moult apparently got evidence from nuclear magnetic resonance spectroscopy that the AlphaFold2's software was not predicting very well, but he seems to have disregarded that evidence, by declaring the software a monumental success. This may be the bias that can arise when someone is yearning to have a "very special moment" of triumphal celebration.
The Nature article makes it rather clear that the CASP14 competition did not use an effective blinding protocol. Quoting a person (Andrei Lupas) who is listed as the person responsible for assessing high accuracy modeling for the CASP14 competition, the article states this:
"AlphaFold’s predictions arrived under the name 'group 427', but the startling accuracy of many of its entries made them stand out, says Lupas. 'I had guessed it was AlphaFold. Most people had,' he says."
This confession makes it sound like a simplistic blinding protocol was used, allowing the judges to guess that "group 427" was the AlphaFold2 software of the prestigious DeepMind corporation, the company that won the previous CASP competition. So the clumsy attempt at blinding didn't work well. We can only guess how much the "prediction success" analysis was biased once people started thinking that the "group 427" (i.e. AlphaFold2) was supposed to win higher scores. Any moderately skilled computer programmer could have easily figured out a double-level blinding system that would have avoided such a problem, one in which a unique name was used for the source of each prediction. Since the CASP14 competition apparently didn't do blinding effectively, what confidence can we have that it did security correctly? If the security wasn't done correctly, that would be a reason for lacking confidence in the predictive results; for one or more competitors might have not been blind about the 3D shapes they were trying to predict.
AlphaFold2 used frequentist inference when it used template-based modeling to get its best results. I can give another example to explain how such frequentist inference can give "blind predictions" that involve no real understanding. I might be given some data on some death cause described only as "Death Cause #523." The data might include how many people of different ages in different locations died with this cause. Using frequentist inference and some "deep learning" computer program, I might be able to predict pretty well how many people of different ages in different places will die next year from this cause. But I would have no understanding of what this death cause was, or how it killed people. That's how it so often is with frequentist inference or "deep learning." You can get it to work fairly well without understanding anything about causes.
I can sketch out how it might actually look like if humans were to solve the protein folding problem:
(1) There would be a software for protein shape prediction, and that software would have all of its code published online.
(2) The software would not use any "deep learning" database or "template library" database unavailable to an organism, some database derived from studying countless different organisms. When doing a prediction for a protein in one particular organism, the software would only use the information in the organism's genome.
(3) The software would only make predictions based on only the information in an organism and known principles of chemistry, physics and biology.
(4) The software would be very well-documented internally, so that each step of its logic would be justified, and we could tell that it was using only principles of nature that have been discovered, or information existing in the organism having a particular protein.
(5) A user of the software could select any gene in the human genome (or some other genome) to test the software.
(6) The software would then generate a 3D model of a protein using the selected gene, without doing any very lengthy number crunching unlike anything that could take place in an organism.
(7) A user could then compare the accuracy of the resulting model, by comparing a visualization of the software's 3D model with a visualization previously produced by some independent party and stored in one of the protein databases.
(8) If automated techniques were used to judge accuracy, only the more accurate GDT_HA method would be used, rather than the less accurate GDT_TS method.
(9) You would be able to test the software using genes of greater-than-average-complexity, and on proteins that require chaperone molecules for their folding, and the predictive results would be very accurate.
The situation with the AlphaFold2 software is light-years short of the type of situation I have described. The protein folding problem has not at all been solved, because it is still the case that no can explain how it is that mere polypeptide chains (sequences of amino acids) tend with such reliable regularity and amazing speed to form into functionally useful 3D protein shapes. The article here discusses why protein folding involves enormous fine-tuned complexity and interlocking dependencies of a type that would seem to make it forever impossible to very reliably predict the more complex protein shapes from the mere amino acid sequences in a single gene. We read the following:
"Protein folding is a constantly ongoing, complicated biological opera itself, with a huge cast of performers, an intricate plot, and dramatic denouements when things go awry. In the packed, busy confines of a living cell, hundreds of chaperone proteins vigilantly monitor and control protein folding. From the moment proteins are generated in and then exit the ribosome until their demise by degradation, chaperones act like helicopter parents, jumping in at the first signs of bad behavior to nip misfolding in the bud or to sequester problematically folded proteins before their aggregation causes disease."
Lior Pachter (a CalTech professor of computational biology) states that while the AlphaFold2 software has made significant progress, "protein folding is not a solved problem." He scolds his colleagues who have got sucked in by the hype on this matter.
In an article in Chemistry World, we read the following:
"Others take issue with the notion that the method ‘solves the protein folding problem’ at all. Since the pioneering work of Christian Anfinsen in the 1950s, it has been known that unravelled (denatured) protein molecules may regain their ‘native’ conformation spontaneously, implying that the peptide sequence alone encodes the rules for correct folding. The challenge was to find those rules and predict the folding path. AlphaFold has not done this. It says nothing about the mechanism of folding, but just predicts the structure using standard machine learning. It finds correlations between sequence and structure by being trained on the 170,000 or so known structures in the Protein Data Base: the algorithm doesn’t so much solve the protein-folding problem as evade it. How it ‘reasons’ from sequence to structure remains a black box. If some see this as cheating, that doesn’t much matter for practical purposes."
No doubt, the fine work of the DeepMind corporation in improving the performance of their AlphaFold2 software will prove useful somehow, in areas such as the pharmaceutical industry. But when a modest success was achieved, that should not be inaccurately described as a solution to one of nature's great mysteries.




No comments:
Post a Comment