According to Darwinist materialism, your mind is merely the product of your brain (or maybe the same thing as your brain states). Also according to materialism, your physical body arose purely because of natural factors such as low-level molecular interactions, made possible by accidental random mutations in DNA occurring long ago. There are many ways of debunking such ideas. Seven of these ways have one thing common: they all involve unsolved questions of navigation, mainly problems of how a body could find the right spot to do something.
Navigation Problem #1: How Could a Brain Select One Particular Spot for a Memory to be Stored, and Direct Sensory Information to Be Stored at That One Spot?
When information is written, either (1) writing occurs at some "current writing position" or (2) some particular spot is chosen for the memory to be written. If you have a conscious agent involved, choosing such a spot for the information to be written is a simple matter. For example, if you are a student taking notes in a class, you may have a notebook with pages. Each time you want to write something, you follow a simple algorithm such as "Keep writing after the last line where something was written, and if nothing is written on the notebook, start writing on the first page."
Choosing a particular spot where information should be written is also easy for a machine such as a computer. Such a machine has something called an operating system, a collection of software subroutines used for routine system tasks such as storing information in a new file. Somewhere in that operating system is some subroutine that is used when you store a new file to your computer. Maybe the subroutine involves some logic such as "(1) find out the length of the data to be stored, in bytes (2) look for the first blank unused area on a hard disk that has a size equal to or greater the data length; (3) write the file at that location."
Some systems are constructed in a way so that there is a kind of "current writing position," so no choice needs to occur about where to write the information. For example, consider a video cassette recorder such as was used in the 1990's. The tape in such a cassette would be threaded through the machine, with some particular spot on the tape (which we can call the "current writing/reading position") being adjacent to the read/write head of the machine. If you pressed the Record button, the recording (a storage of new information) would take place at whatever spot was that "current writing/reading position" in the video cassette.
But there are some big problems involved in any theory that sensory information is stored in some memory storage spot in a brain. Specifically:
(1) When people learn things, they do not at all consciously select some particular spot in the brain where the new information should be written.
(2) The brain has nothing at all corresponding to the operating system of a computer, so we cannot imagine a memory storage spot being selected by something like an operating system subroutine.
(3) The brain has no physical structure with anything like a "current writing position" or a "current reading position." There is a nothing in a brain anything like a read/write head in a VCR or a computer.
We therefore have a great explanatory problem: if a brain were to store a new memory in some particular spot in the brain, why would the brain have navigated to that particular spot to store the memory? There would seem to be no credible answer to this problem. The problem casts great doubt on all claims that the brain stores human memories.
You do not get around this problem by speculating that maybe new memories are stored in multiple places. Under such a scenario, you have the same problem. If you don't have a scenario that can explain why a brain would have chosen Neural Location #342464 to store a new memory out of a billion neural locations, you don't solve such a problem by turning it into a problem why a brain would have chosen Neural Location #342464, Neural Location #15342464, and Neural Location #6344464 out of a billion neural locations.
You do not get around this problem by speculating that a new memory is first formed in the hippocampus and then migrates to the cortex, a tall tale told by many neuroscientists without any good evidence to back it up. Scientists tell this story because the hippocampus (wrongly claimed as a site of memory formation) is a particularly unstable area, with its dendritic spines having a particularly short lifetime. This study found that dendritic spines in the hippocampus last for only about 30 days. This study found that dendritic spines in the hippocampus have a turnover of about 40% each 4 days. Because it is typically claimed that memories are stored in synapses, and synapses are attached to such dendritic spines, scientists have created the tale of memories migrating from the particularly unstable hippocampus to the cortex (ignoring the fact that dendritic spines still don't last for years in the cortex). If you believe that tale, the problem I am discussing here simply becomes twice as bad; for now you have both the problem of why a memory would be stored in some particular millionth of the hippocampus, and also why some particular millionth of the cortex would be chosen as the place to which this memory would migrate.
The problem discussed here is described more fully in my post "No One Can Credibly Explain Why a Brain Would Store a Memory in One Specific Spot." The problem would not exist if the brain had some structure resembling a stack of pages. Then we could simply believe that the brain creates a new page at the top of such a stack whenever a new memory is formed. But the brain has no structure bearing any resemblance to a stack of pages. The problem would not exist if the brain had some structure resembling a cursor or current writing position. But the brain has no such structure. The problem discussed in one of very many reasons for doubting that your brain is the storage place of your memories, and for thinking that memory must be a spiritual facility, not a neural one.
Navigation Problem #2: How Could a Brain Instantly Find One Particular Spot Where the Right Memory Was Stored?
Humans routinely display the ability to instantly recall learned information, given a name, date or image. So, for example, if you say "death of Lincoln," I will instantly be able to recite various facts about the death of Abraham Lincoln, such as that it occurred because John Wilkes Booth shot Lincoln through the back of his head at Ford's Theater on April 15, 1865. If we believe that a memory is stored in some tiny little spot in the brain, such as storage spot 186,395 out of 950,000, then we have the problem: how was the brain able to instantly find that exact tiny spot where the memory was formed? This difficulty is a "show stopper" for all claims that a memory is stored in one exact spot of a brain, an insuperable difficulty.
We cannot get around such a difficulty by imagining that a brain uses the type of things that a book or a computer use to allow instant retrieval. Books and computers use information addressing, sorting and indexes to allow instant access of a particular data item. The brain has neither addressing nor indexes nor sorting. Unlike houses that have street addresses, neurons don't have neuron numbers or any other addressing system. Storing a memory in a brain would be like throwing a little 3" by 5" card into a giant swimming pool filled to the top with a million little 3" by 5" cards. Just as it should take you a very long time to find a specific piece of information stored in such a swimming pool, it would take you a very long time to find in the brain some particular piece of learned information, if it was stored in one tiny spot, like a book stored in one spot on the shelves of a huge library.
You do not at all get around this difficulty by suggesting the idea that a memory or a piece of learned information is scattered or distributed in multiple locations across the brain. The main difficulty is explaining instantaneous recall. If a brain has to search scattered storage locations in the brain, that would not be any easier than finding a single storage location; it would instead be harder. We would then have the same problem: how is it that those exact locations can instantly be found? Similarly, if a family is somewhere in New York City, and you don't know their address, without an electronic device you won't be able to find the family very quickly; and it's not going to be any easier if the family is scattered across three different apartments in different parts of the city, which would make finding the family even harder. You do not solve a "how was the needle instantly found in the haystack" problem by converting it to the even harder problem of "how were just the right few needles instantly found in multiple haystacks?" Moreover, the idea of a brain instantly bringing together scattered fragments to instantly make a unified conceptual whole creates an "instant reassembly" problem that would be an additional explanatory nightmare, with such a thing being some miracle of instant assembly as implausible as someone instantly assembling cut-up pieces of a photo after the pieces had been scattered in pages of different books on different bookshelves.
This "speed of human recall" problem becomes much worse when we consider that brain signals have an average transmission speed much slower than the "100 meters per second" figure that is commonly given (which is the fastest speed that any nerve signal can travel over any part of the brain) A typical brain signal traveling from one part of a brain to another would have to pass across many chemical synapses, and each time that happens there would be a delay. The effect of cumulative synaptic delays would mean that brain signals must typically travel from one area of a brain to another at a sluggish speed of something like about one centimeter per second or less. Even if a brain somehow knew exactly where to find some information it needed, the retrieval of such information would be too slow to explain instant human recall.
The ability of humans to instantly recall relevant information after hearing a single word is one of many strong reasons for rejecting the claim that memories are stored in the human brain. We do not recall at the speed of brains. We recall at the speed of souls.
Navigation Problem #3: How Could a Cell Find the Exact Spot in DNA Where There Existed Some Gene the Cell Needed to Read to Construct a Protein?
The short average lifetimes of synapse proteins (less than two weeks) is one of the main reasons for rejecting the neuroscientist dogma that memories are stored in synapses (such a lifetime being only about a thousandth of the longest length that humans can remember things). Cells are constantly creating new proteins to replace proteins that disappeared because of the short lifetimes of proteins. The page here has a chart showing the lifetimes of human proteins, and we see a bar graph showing most of the proteins have a half-life between about 10 hours and 70 hours. A muscle protein might live for three weeks, but a liver protein might live for only a few days. To create new proteins, a cell uses a process called gene transcription. In this process a particular gene in DNA will be converted to a messenger RNA molecule that helps to build the new protein.
Cell transcription occurs quickly. The source here lists a time of ten minutes for a gene to be transcribed by a mammal, but another source lists a speed of only about a minute. The great majority of that is used up by the reading of base pairs from the gene, with typically more than a 1000 base pairs being read each time a gene is transcribed. The finding of the correct gene to read in DNA seems to occur in only seconds, not minutes, or at most a few minutes.
Descriptions of DNA transcription fail to explain a huge issue: how does a cell find the right gene in DNA so quickly? Human DNA contains more than 20,000 genes, each of which is just a section of the DNA. The DNA is like an extremely long necklace of many thousands of beads, and a typical gene is like a group of several hundred of those beads. We should actually imagine multiple such necklaces, because DNA is scattered across 23 different chromosome pairs. Now if genes had gene numbers, and DNA was a set of numbered genes in numerical order, it might be easy to find a particular gene. So if a cell knew that it was trying to find gene number 4,233, it could use a binary search method that would allow it to find that gene pretty quickly. Such a method might sound like Bob using a binary search method efficiently in the dialog below:
Jane: Okay, I picked a date in world history. Try to guess it.
Bob: Was it after the first century AD?
Jane: Yes.
Bob: Was it in the past thousand years?
Jane: Yes
Bob: Was it in the past 500 years?
Jane: No.
Bob: Was it between 1250 and 1500?
Jane: Yes.
Bob: Was it between 1375 and 1500?
Jane: Yes.
Using such a binary search method, Bob will find the correct year within several more guesses.
But no such method can be used within the human body. Genes do not have gene numbers that can be accessed within the human body, and DNA is not numerically sorted. DNA has no indexes that might allow a cell to find some particular gene that it was trying to find within DNA. So we have an explanatory "needle in a haystack" problem. Or we might call it a "needle in the haystacks" problem, because human DNA is scattered across 23 different chromosome pairs, as shown in the diagram below:
A scientific text tells us some information that makes this explanatory problem seem more pressing:
"One might have predicted that the information present in genomes would be arranged in an orderly fashion, resembling a dictionary or a telephone directory. Although the genomes of some bacteria seem fairly well organized, the genomes of most multicellular organisms, such as our Drosophila example, are surprisingly disorderly. Small bits of coding DNA (that is, DNA that codes for protein) are interspersed with large blocks of seemingly meaningless DNA. Some sections of the genome contain many genes and others lack genes altogether. Proteins that work closely with one another in the cell often have their genes located on different chromosomes, and adjacent genes typically encode proteins that have little to do with each other in the cell. Decoding genomes is therefore no simple matter. Even with the aid of powerful computers, it is still difficult for researchers to locate definitively the beginning and end of genes in the DNA sequences of complex genomes, much less to predict when each gene is expressed in the life of the organism. Although the DNA sequence of the human genome is known, it will probably take at least a decade for humans to identify every gene and determine the precise amino acid sequence of the protein it produces. Yet the cells in our body do this thousands of times a second."
We have here a very severe navigation problem. A cell is somehow able to find the right gene in only seconds or a few minutes when a new protein is made, even though DNA and chromosomes seem to have no physical organization that could allow for such blazing fast access to the right information. In an article on Chemistry World, we read this:
"How does the machinery that turns genes into proteins know which part of the genome to read in any given cell type? ‘To me that is one of the most fundamental questions in biology,’ says biochemist Robert Tjian of the University of California at Berkeley in the US: ‘How does a cell know what it is supposed to be?"
Biochemist Tjian has spoken just as if he had no idea how it is that a cell is able to navigate to the right place to read a particular gene in DNA. Later in the article we read this:
"For one thing, the regulatory machinery ‘is unbelievably complex’, says Tjian, comprising perhaps 60–100 proteins – mostly of a class called transcription factors (TFs) – that have to interact before anything happens. ....As well as promoters, mammalian genes are controlled by DNA segments called enhancers. Some proteins bind to the promoter site, others bind to the enhancer, and they have to communicate. ‘This is where things get bizarre, because the enhancer can sit miles away from the promoter,’ says Tjian – meaning, perhaps, millions of base pairs away, maybe with a whole gene or two in between. And the transcription machinery can’t just track along the DNA until it hits the enhancer, because the track is blocked. In eukaryotes, almost all of the genome is, at any given moment, packaged away by being wrapped around disk-shaped proteins called histones. These, says Tjian, ‘are like big boulders on the track’: you can’t get past them easily.... ‘Even after 40 years of studying this stuff, I don’t think we have a clear idea of how that looping happens,’ says Tjian. Until recently, the general idea was that the TFs and other components all fit together into a kind of jigsaw, via molecular recognition, that will bridge and bind a loop in place while transcription happens. ‘We molecular biologists love to draw nice model schemes of how TFs find their target genes and how enhancers can regulate promoters located millions of base pairs away,’ says Ralph Stadhouders of the Erasmus University Medical Centre in Rotterdam, the Netherlands. ‘But exactly how this is achieved in a timely and highly specific manner is still very much a mystery.’ "
Later in the article Tjian says he was shocked by the speed at which some of the process occurs. He expected it would take hours, but found something much different:
"The residence times of these proteins in vivo was not minutes or hours, but about six seconds!’, he says. ‘I was so shocked that it took me months to come to grips with my own data. How could a low-concentration protein ever get together with all its partners to trigger expression of a gene, when everything is moving at this unbelievably rapid pace?’ "
The rest of the article is just some speculation, which Tjian mostly knocks down, and the article itself calls "hand-wavy." We are left with the impression that no one understands how cells are able to instantly find the right gene.
On page 100 of the very interesting work "Theory of Directed Evolution" scientist Alexey V. Melkikh asks this:
"How does a protein during genome regulation find its only place on the DNA molecule? If it is based on the key-lock principle, how does the protein not confuse its binding site with someone else's? How many erroneous attempts at linking should it make until it finds its site? Why does it not get stuck in someone else's deep potential hole? All these processes should drastically reduce the efficiency of genome regulation."
The question I raise in this section of this post is a question raised, but never answered, by the latest (32nd) edition of Harper's Illustrated Biochemistry, which states this:
"The question 'How does RNAP [RNA polymerase] find the correct site to initiate transcription?' is not trivial when the complexity of the genome is considered....The situation is even more complex in humans, where as many as 150,000 distinct transcriptions sites are distributed throughout [three billion base pairs] of DNA."
The textbook gives us a very detailed discussion of things such as promoters, but the discussion fails to answer the question of how this "finding the needle in a haystack" could occur so quickly.
The realities discussed above suggest two credible alternatives, both of which are incompatible with materialism and ideas that biological organisms arose accidentally:
(1) It could be that cells have some fantastically complex machinery or engineering, mostly undiscovered, allowing cells to find just the right gene almost instantly. This would have to be some miracle of complex design gigantically more impressive than the anatomy of an eye, some accidentally unachievable organization that would strongly discredit claims of accidental human origins.
(2) Or, there may be some astonishing organizational reality similar to what I have described as GOAL (Global Organizing Activity of a Life Force), causing cells to find "needle in a haystack" targets instantly, something inexplicable through molecular biology, in addition to other purposeful teleological effects.
The problem discussed here is a problem similar to Navigation Problem #2 above. In both cases materialists would have us believe in some "instantly finding the needle in a haystack" effect that should be impossible under the assumptions of materialists. The main difference is that with Navigation Problem #2 we can dispose of the problem entirely by discarding the unfounded notion that memories are stored in brains, an idea never justified partly because the microscopic examination of brain tissue never supported it (human memories have never been found through microscopic examination). But with the problem of how a cell is able to almost instantly find the right gene, it's a different type of situation, because scientists have determined through microscopic examination that genes (small parts of DNA) really do store the amino acid sequences of proteins.
Navigation Problem #4: How Could the Proteins of Protein Complexes Ever Find Their "Team Members" to Create the Protein Complexes?
Most of the more than 20,000 types of proteins in the human body do not work in a standalone way, but as team members of larger structures that are called protein complexes. A protein complex will consist of multiple proteins attached together so that some particular function can be performed. A rarely considered question of great importance is: how do such protein complexes form? You cannot answer this question by referring to DNA, which does not specify which proteins belong to particular protein complexes.
There is a factor that makes the formation of protein complexes very hard to explain: the fact that there exists so many thousands of types of proteins in the human body. It is relatively easy to explain the formation of something when there are only a few types of things that might combine together. Consider the formation of water in interstellar space. There are not very many types of gases floating around in interstellar space, so it is not so improbable that two hydrogen atoms and one oxygen atom might combine to form a water molecule consisting of two hydrogen atoms and one oxygen atom. But within the body there are more than 20,000 genes that can specify the amino sequence of more than 20,000 types of protein molecules. A particular protein complex requires very specific proteins to combine together to form that specific protein complex, just as a sentence requires very specific words to join together in a particular way to have the semantic content of the sentence. Given that the number of protein types in the human body is very roughly about the same as the number of words in a particular language, and given that the number of types of proteins in a protein complex is about the same as the number of words in an average sentence, it seems that each time that a useful protein complex forms we would seem to require luck at least as great as would be required for someone to form a useful sentence by about 15 times randomly opening a page of a dictionary, and picking the first or last word on that page. The chance of doing such a thing successfully is incredibly low.
Could it be that the unlikelihood of the chance formation of a useful protein complex is not so small, because maybe all of the proteins used in the protein complex are all specified in contiguous spots in DNA, and that such a thing makes it more likely that the protein complex may form? Not actually, because a very important and little-known fact is that the proteins used in protein complexes are scattered in diverse spots of different chromosomes. So finding the right proteins to make up a particular protein complex would be like finding ten needles scattered in ten haystacks.
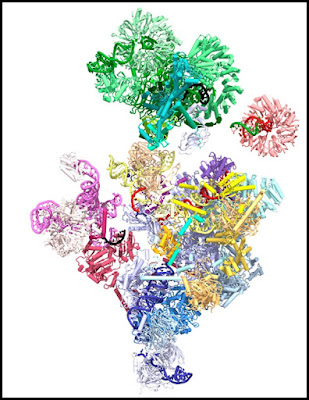
I can give some examples to back up this claim. Below is a depiction of the spliceosome protein complex, with different colors being different proteins:
At the site here, we read this about the human spliceosome:
"The spliceosome is a complicated and formidable example of a multi-subunit molecular machine, with the pre-catalytic form being the largest spliceosomal complex, containing 5 RNA molecules and 65 proteins, in addition to a substrate mRNA precursor. The arrangement and activities of all of these has to be intricately coordinated, paradoxically to catalyse a rather simple chemical reaction."
The paper here describes the spliceosome as a highly dynamic machine, like some race car that has its parts changed or replaced by a pit crew as the race car stops for pit stops:
"Indeed, ∼45 proteins are recruited to the human spliceosome as part of the spliceosomal snRNPs, whereas non-snRNP proteins comprise the remainder. The composition of the spliceosome is highly dynamic with a remarkable exchange of proteins from one stage of splicing to the next. These changes are also accompanied by extensive remodeling of the snRNPs within the spliceosome."
Here are some proteins used by the protein complex known as the spliceosome:
Protein | Number of amino acids | Location of Its Gene |
| 793 | On Chromosome 22 | |
| 462 | On Chromosome 19 | |
| 501 | On Chromosome 1 | |
| 683 | On Chromosome 1 | |
522 | On Chromosome 9 | |
2335 | On Chromosome 17 | |
972 | On Chromosome 17 | |
| 499 | On Chromosome 19 | |
522 | On Chromosome 9 | |
| 800 | On Chromosome 11 | |
| 586 | On Chromosome 5 | |
| 420 | On Chromosome 5 | |
112 | On Chromosome 19 | |
855 | On Chromosome 19 | |
758 | On Chromosome 19 | |
514 | On Chromosome 4 |
We see that the proteins needed to make the spliceosome complex are not at all located in some contiguous spot, but are scattered across many different chromosomes. I can give another example of such a thing. Below are some of the proteins used in the RNA Polymerase III protein complex.
Protein | Number of amino acids | Location of Its Gene |
346 | On Chromosome 6 | |
132 | On Chromosome 13 | |
534 | On Chromosome 1 | |
398 | On Chromosome 8 | |
708 | On Chromosome 16 | |
223 | On Chromosome 5 | |
204 | On Chromosome 22 | |
148 | On Chromosome 7 | |
108 | On Chromosome 16 | |
210 | On Chromosome 19 | |
| 127 | On Chromosome 22 | |
150 | On Chromosome 3 | |
54 | On Chromosome 8 | |
67 | On Chromosome 11 | |
We see that the proteins needed to make the RNA polymerase III protein complex are not at all located in some contiguous spot, but are scattered across many different chromosomes. I can give another example of such a thing. Below are some of the proteins used in the human 26S proteasome protein complex (which according to the paper here consists of 31 protein subunits):
Protein | Number of amino acids | Location of Its Gene |
| 241 | On Chromosome 6 | |
| 201 | On Chromosome 1 | |
| 205 | On Chromosome 17 | |
| 264 | On Chromosome 1 | |
| 263 | On Chromosome 14 | |
| 239 | On Chromosome 17 | |
| 248 | On Chromosome 20 | |
| 263 | On Chromosome 11 | |
| 234 | On Chromosome 7 | |
| 255 | On Chromosome 14 | |
| 261 | On Chromosome 15 | |
| 241 | On Chromosome 1 | |
| 243 | On Chromosome 14 | |
| 248 | On Chromosome 20 | |
We see that the proteins needed to make the human 26S proteasome protein protein complex are not at all located in some contiguous spot, but are scattered across many different chromosomes.
It seems that we have no basis at all for thinking that some contiguous placement in DNA can reduce the prohibitive probabilities calculated above. We are left with the conclusion that every time that one of the larger protein complexes forms, we are seeing a system assembly that should be accidentally unachievable.
A scientific paper states this: "While the occurrence of multiprotein assemblies is ubiquitous, the understanding of pathways that dictate the formation of quaternary structure remains enigmatic." In other words, scientists don't understand how protein complexes are able to form. Similarly, the scientific paper here states the following:
"The majority of cellular proteins function as subunits in larger protein complexes. However, very little is known about how protein complexes form in vivo.“
Navigation Problem #5: How Could Proteins in a Cell Find Where to Go to in the Cell?
A scientific article entitled "Mysteries of the Cell" raises some profound questions. One of these is: how does a cell know how to stop growing? We read this:
"Within specific cell types, cells actually stick to 'a fairly narrow range of sizes,' says cell biologist Marc Kirschner of Harvard Medical School in Boston. Biologists have puzzled over how cells know when they’ve reached the right size. 'The question has been there for a long time. The answers haven’t,' Kirschner says."
The article raises the question: how does the cell position its proteins? You may realize how puzzling this question after you remember that cell diagrams are almost all very misleading visuals which depict cells as a thousand times simpler than they are. A cell diagram will show 20 or 30 organelles in a cell, but the actual number is typically more than 1000. A cell diagram will typically depict a cell as having only a few mitochondria, but cells typically have many thousands of mitochondria, as many as a million. A cell diagram will typically depict a cell as having only a few lysosomes, but cells typically have hundreds of lysosomes. A cell diagram will typically depict one or a few stacks of a Golgi apparatus, each with only a few cisternae, but a cell will typically have between 10 and 20 stacks, each having as many as 60 cisternae. There are about 200 different types of cells in the human body, and in each type, there are hundreds or thousands of types of proteins that must each go to the right place. How do such proteins know how to go to the right place in a cell?
The article tells us this:
"The latest analyses suggest that some of our cells make more than 10,000 different proteins. And a typical mammalian cell will contain more than a billion individual protein molecules. Somehow, a cell must get all its proteins to their correct destinations—and equally important, keep these molecules out of the wrong places...Biologists stress that the mystery of how cells place their protein repertoire is far from solved."
The article reminds us that it is extremely fallacious to think of a cell as some disordered bag of proteins. A cell is more like a factory consisting of many different machines: the many types of organelles within the cell. Each protein is like one of the parts of the machines in a factory. Just as almost all the parts of a factory's machines must end up in the right place for the factory to work right, almost all the proteins of a cell must end up in the right organelles. DNA does not specify where particular proteins should go in a cell.
By doing a Google search for "protein localization" you can find some sites that discuss some fantastically intricate mechanisms by which proteins supposedly are delivered to the right organelles within cells. For example, you can look at the Slideplayer.net slideshow here for an overview. We read about a fantastically intricate mechanism involving extraordinarily complex machinery involving many different proteins, just as an explanation for how a protein can end up in the right place in the cell. The page here gives us a taste of how fantastically complicated is the molecular machinery allowing protein molecules to find the right places in cells:
"The protein distribution among cellular compartments depends on four main factors: (1) the number and types of localization protein signals, (2) the concentration of freely diffusing molecules, (3) the relative strength of each signal, and (4) the activity and concentration of localization signal receptors...Modifying the binding affinity of the localization signal for protein corresponding receptor is one of the most common methods by which cell controls the distribution of protein. Post-translational modification of the cargo protein in or near the localization signal achieves this affinity modulation. This type of mechanism usually involves serine/threonine phosphorylation, however may also occur through lysine sumoylation, tyrosine phosphorylation, or lysine acetylation. These modifications interfere with localization receptor binding. Signal addition is another common method of adjusting protein localization....Protein sequestration within a compartment happens if the number of binding sites within the compartment remarkably decreases the protein mobility so the export rate from the compartment is decreased. Tethering, or anchoring, is a unique form of sequestration where the protein is embedded in or bound to an essentially immobile component with respect to other components, like the chromatin, cytoskeleton, or plasma membrane. Tethering decreases the soluble pool available to be acted on by transport receptors."
Delving into this topic of protein localization will tend to strengthen a person's suspicion that proteins are absolutely inexplicable as results of random mutations. A study of the subject will make you suspect:
(1) that protein molecules are very sensitive-to-small-changes kind of molecules that would never be both functional and arriving at the right targets if such molecules had only half of their parts;
(2) that the successful positioning of proteins within cells requires a fantastically intricate teamwork more impressive than the kind of teamwork needed to produce a hit movie or a hit Broadway musical.
Experiments on small changes in protein molecules show that they are very sensitive and will typically be broken and dysfunctional by only small changes. A paper describing a database of protein mutations tells us that "two thirds of mutations within the database are destabilising." Generically we can say that not only does a protein molecule have to have almost all of its amino acid sequence and just the right shape, but also that protein molecules require a fine-tuned supporting infrastructure of extreme complexity, just so that the protein molecule ends up in the right place in the cell where it can do some good.
This problem differs from Problem #2. Scientists have written very much on the topic of protein localization. They seem to have put together an extremely complicated story-line that may well be a partial explanation of how proteins are able to get to the right locations in cells, although we are left with a suspicion that the full story is ten times or a hundred times more complicated. Cell biologists have not ignored the problem of protein localization. But it's a very different situation with Problem #2, the problem of how a brain could instantly find just the right information to allow someone to instantly answer a question. Neuroscientists have no credible answer to give us on this question, and don't even have the beginning of an answer. The problem is that we know (from the devices humans have made) some of the things that allow instant retrieval of information (things like sorting, addressing and indexing), and the brain has no such things.
Navigation Problem #6: How Could a Cell Find the Exact Spot Where It Should Go to in a Developing Human Body?
Another great unsolved navigation problem is: how could newly formed cells in a developing human body navigate to find the right places to go to so that extremely complex human anatomical structures are formed? Consider, for example, a human vision system, consisting of the human eye, the optic nerve, and parts of the brain relating to vision perception. The human vision system is built from many different types of cells, each of which has to end up in the right place for an eye to function properly. Or consider the human circulatory system, consisting of the lungs, the heart, veins and arteries. Such a system requires many different cells, which each have to end up in the right places for the system to function properly.
DNA does not specify the structure of cells, and does not at all specify where particular cells should go in a body. Other than making incorrect insinuations suggesting DNA tells cells where to go, when he or she is asked the question of how cells are able to find the right place to go to in the body, a developmental biologist has nothing to offer except an extremely far-fetched story that does not hold up to critical scrutiny. The typical answer given will make some reference to "morphogen gradients," referring to some paper written by biologist Lewis Wolpert. We will often be told that Wolpert introduced something called the "French flag model" that helps explain how cells find the right positions, after being instructed where to go to by "morphogen gradient" chemical signals. There is no robust evidence behind such a claim, and there is no strong reason to believe that morphogen gradients do anything substantive to solve the great problem of how cells are able to find the right positions in a developing human body.
Let me summarize some of what I discuss at greater length in my post "A Critique of Morphogen Gradients, a Tall Tale of Developmental Biologists."
(1) In 1969 Lewis Wolpert wrote a paper entitled "Positional Information and the Spatial Pattern of Cellular Differentiation," raising a problem he called a "French flag problem" involving how cells could find the right positions. He described various hypothetical ways in which chemical signals might cause a cell to move to a particular spot on a flat two-dimensional space. He confessed on page 22 that "what is completely lacking is a model of how such gradients are established, regulated, or exert their effect, except in the vaguest of terms." He did not refer to a "French flag model" or claim to have any good model of how cells could find the right position in bodies.
(2) The term "French flag model" was almost never used in the scientific literature until about 1998. Then between 1998 and the year 2010 more and more scientific papers started to refer to a "French flag model," with about 60 papers a year referring to such a thing by the year 2010. A myth had been socially constructed, that a scientist (Wolpert) had advanced some "French flag model" when his paper never even used such a term. This is the same Wolpert who confessed in 2015 (in the quote below) that "we still don't have a molecular basis for it." Below we see the Google NGram Viewer showing how references to a "French flag problem" started changing to references to a "French flag model," even though no actual model had been introduced.
(3) For years many developmental biologists eagerly went looking for evidence of the imagined "morphogen gradients" imagined as signals of positional information. They only found a small number of cases they could claim as evidence for such things, with the results being no greater than you would expect given a large number of developmental biologists eagerly seeking such an effect, rather like believers that clouds can be animal ghosts eagerly scanning the skies for cloud shapes looking like animals. All evidence found thus far for "morphogen gradients" can be plausibly explained as mere examples of pareidolia, like cases of someone eagerly scanning his toast every morning for something that looks like the face of Jesus, and eventually reporting some result that pleases him.
(4) Lewis Wolpert himself confessed in 2017 how little evidence there was for the idea of these "morphogen gradients." He stated in 2017, "It is still not clear whether diffusible morphogens provide cells with positional information and so pattern a tissue during development." Another paper ("Biological notion of positional information/value in morphogenesis theory") states this: "The fundamental role of morphogens as a basis for positional information in a complicated living body is still questionable." Another paper states that "arguments against morphogen movement by diffusion have been raised by many." In 2015 Lewis Wolpert was asked this: "Where do you think the French Flag model fits with our current understanding of positional information, and what do you think are the exciting questions at the moment?" Responding to this question about the theory of morphogen gradients providing positional information to cells, Wolpert answered this:
"There are problems we haven’t solved. It is terrible, but we still don’t have a molecular basis for it. If I still had an active lab, finding the molecular basis for positional information would be my objective, but would be quite tricky, since I’m not a biochemist or molecular biologist. There is one case of a molecule that might encode positional information, Prod 1, which is graded along the amphibian limb and was discovered by Jeremy Brockes. But it would be nice to find similar molecules in other systems."
We can compare the theory of morphogen gradients to the theory of panspermia, the theory that life on Earth came from outer space. Panspermia does not solve the problem of how life arose, because it leaves you with this unanswered problem: how could life have originated in outer space? That's just as hard as the problem of how life could have originated on Earth, so there's no real explanatory progress made if you imagine life came from outer space. Similarly, the theory of morphogen gradients attempts to solve the problem of how cells find their correct position in a forming human body by imagining that their correct position is told them by signals from some external chemicals (morphogen gradients). But that leaves unanswered the equally big question: how could such chemicals know what the correct positions of the cells should be? They could not have got the information from DNA or genes, which specify neither how to make cells nor how cells should be arranged. So imagining signaling chemicals that tell cells where to go accomplishes nothing, because no explanation is given as to how such chemicals knew where the cells should go. We've simply gone from "inexplicably high-knowledge cells" to "inexplicably high-knowledge signaling chemicals," which accomplishes nothing in reducing the mystery of how a fertilized ovum progresses to become an adult organism.
Until the student asked this.
Navigation Problem #7: How Could a Brain Ever Recognize Quickly Enough for You to Navigate Quickly in Your Neighborhood?
Let us consider a simple case of a person navigating around in his city neighborhood, without looking at any street signs. Someone may run from his home to a grocery story seven blocks away. This requires all kind of wondrous mental work for it to occur. Each time the person comes to the end of some block, he must instantly get some idea about where he is, and which direction to travel in next. But this cannot occur by the person making an exact match between the spot he is at and what he sees around him, because there is no exact match between what he sees and something he has seen before. For one thing, the person's position is slightly different each time. For another thing, the street will look a little different time each time he sees it, with different cars parked on the street, visual differences caused by time of day and weather, and different people on the street. What must go on is that each time the person comes to the end of a block, he must do a pattern matching, rather like the pattern matching when he identifies some celebrity in a new photo, even though he may have never seen that celebrity looking just exactly that way before.
No one has a credible explanation of how a brain could do pattern matching. There is no good evidence that it does such a thing, and we merely know that human minds do pattern matching. Claims that a fusiform area of the brain activate more highly during face recognition are not well supported, and are backed up by only studies involving too-small study group sizes and unimpressive signal changes such as about 1%. Discussing something called "face blindness" involving a difficulty recognizing faces, in a recent Harvard Medical article an expert states. "Prosopagnosia, or face blindness, can be caused by a brain injury to occipital or temporal regions, referred to as acquired prosopagnosia, which affects one in 30,000 people in the United States." But brain damage (caused by aging, disease, injuries or brain tumors) is much more common that that, so you would think that if face recognition was done by your brain, that the incidence of acquired prosopagnosia would be much higher than that.
There are computer software and hardware setups that are called neural nets, which can do pattern matching. But that does nothing to establish that a brain could do pattern matching, because these so-called neural nets use a setup that does not match anything that exists in a brain. It has been pointed out that the back propagation algorithm used by computer neural nets should be impossible in a brain. A paper refers to "the dogma, generally accepted for the past 30 years, that the error back-propagation algorithm is too complicated for the brain to implement."
We have in pattern matching an ability of the human mind very different from the instant data recall ability mentioned earlier. This is the ability of a human to do visual instant pattern matching. Just as the physical shortfalls of the brain, severe noise levels of the brain, and severe slowing factors of the brain should make it impossible for you (using only your brain) to ever instantly tell me (using your brain) facts about George Washington as soon as I mention the name of George Washington, similar physical shortfalls of the brain and slowing factors of the brain should make it impossible for you to ever use a brain to quickly navigate around your neighborhood or some other neighborhood.
Pattern matching in a computer's neural net occurs with signals traveling between the nodes of a network at about half the speed of light, with 100% reliability of signal transmission between the nodes of the network. Because of numerous slowing factors such as cumulative synaptic delay and the relatively slow speed of signal transmission across dendrites, average signal transmission through a brain should be slow (on the order of 1 centimeter per second), and also extremely unreliable (because each signal transmission across a chemical synapse occurs with a reliability of only 50% or smaller). But you can recognize a pattern instantly, even when you see something looking different from how you've ever seen it. For example, take a look at the photo below, and try to recognize what it shows.
Now take a look at the image below, and try to recognize what it shows:
It probably took you only a second or so to identify the first image as the the Eiffel Tower in Paris, and the second image as the Taj Mahal in India, even though in both of the distorted images you are seeing these landmarks looking different from any way you have ever seen them. That's how human recognition works. You can recognize things even when they look different from how you've ever seen them, up to a point. So if you have seen bearded fat James Waterson many times, you'll probably recognize him if he comes back from a long trip looking clean shaven and slim.
Your ability to successfully navigate around your neighborhood while you are running, with all kinds of instant recognitions occurring, is something we cannot explain through brain activity. I may note that you do not get around such difficulties by appealing to evolution, and saying that evolution selected for brains that could better recognize safe foods to eat. The issue is that the human brain does not currently have any features that can explain instant recognition. You could only get around such a difficulty by showing that the brain does have such features, not by appealing to some story of what happened in the past. Look at the "Recognition Memory" section of my post here, and you will see that there are not even strong correlates between brain activity differences and acts of human recognition.
Concluding Remarks
In countless ways claims of accidental biological origins and claims that our minds are merely our brains (or the products of our brains) fall flat on their face when we ponder with sufficient care the realities of human minds, the reality of neural physical shortfalls, and the realities of the gigantically complex purposeful targeted molecular and cellular choreography that goes on in human bodies. The realities of human brain shortfalls, the hugely diverse and poorly studied capabilities of human minds and the vast organization and vastly purposeful and blazing fast molecular activity within our bodies collectively scream to us in a very loud voice that such claims are bunk and baloney. The items mentioned in this post are merely like what we see from a boat when looking down into the ocean, with such a visible reality being only a small fraction of a far greater reality of oceanic depth. Properly exploring this oceanic reality would require a deep dive into the topic of anomalous human abilities, a topic our biologists typically refuse to study even a tenth as well as they should, for fear of discovering things that might upset their belief system. You can start to make such a deep dive by reading my 157 posts on paranormal phenomena, or by reading some of my free books here.








No comments:
Post a Comment